Solutions to immunoassay interference, cross reactivity and other challenges
Immunoassays have become invaluable in the generation of data for preclinical and clinical studies, process development/optimization and manufacture of biotherapeutics, and also for new applications such as determining viral titer in cell and gene therapy. Having confidence in the data based on the accurate and precise determination of analytes in complex matrices means overcoming many challenges. These can include interference that reduces robustness, selectivity and specificity, the poor performance of cross-reacting antibody reagents, and time-consuming methods that threaten time plans. Such challenges can be overcome by choosing the right reagents, using the right approach on the right immunoassay platform to deliver reliable data as quickly as possible.
Addressing the complexity of interference
The ideal assay is specific and free from interference, but biological matrices are complex. A recent survey on the use of Ligand Binding Assays (LBA) that we carried out pointed to matrix interference as being the single most important challenge in ligand binding assays for large molecules (72% of respondents).
Sources of interference are varied, depending on the nature of the assay, which may involve the determination of soluble protein biomarkers, free soluble target, or the detection of anti-drug antibodies (ADA). Interference can be defined as, ‘the effect of a substance present in the sample that alters the correct values of the result, usually expressed as concentration or activity, for an analyte’, (1). Interference in immunoassays for the clinical laboratory has been described in detail (2,3) and for therapeutic antibody development in preclinical and clinical studies (4).
Interference should be determined early in method development by assessing parallelism/linearity, by determining the recovery of spiked analyte, and by testing the effect of blocking agents. Interference can also become apparent if inconsistencies in study results become apparent when analyzing the results of a study.
Care should be taken to avoid changes in matrix interference caused by changes in therapy, and sampling techniques should be examined in detail to ensure that they do not result in the release of interfering substances. The specificity of the assay can also be disturbed by isoforms, precursors, related proteins and binding proteins, and some of these effects can be blocked. Endogenous antibodies can interfere by cross-linking or blocking, and these disturbances can be reduced by careful choices of reagents, dilution, depletion or blocking.
The accurate detection of free soluble target can be compromised by dissociation of drug-target complexes during sample preparation and when performing the assay, and this can only be minimized by optimizing reagent concentrations and incubation/assay times. ADA assays are particularly challenging since the drug will always interfere with the assay, and drug tolerance needs to be established during assay development and validation.
Minimal contact times reduce matrix interference
As we have seen, developing and running immunoassays means a balancing act involving many factors, including minimizing interference and maximizing sensitivity. Molecular interactions causing interference in immunoassays are fundamentally a function of affinity, concentration and exposure time. The simplest and most common method to reduce interference is therefore dilution, but this also reduces assay sensitivity. One alternative is to reduce contact times to ensure that the most specific, high-affinity interactions (antibody-antigen) are favored while low affinity interference is minimized. This is possible in a flow-through device that minimizes contact times between reagents, the sample and its matrix.
How to avoid a key threat to immunoassay performance – cross reactivity
Confidence in immunoassay data depends on the accuracy and precision that is only possible from using high quality antibody reagents. This means selecting antibodies with not only high analyte affinity but also specificity, which is the ability to distinguish between the analyte and other structurally similar components. Cross-reactivity, which occurs when an antibody raised against one specific antigen binds to a different antigen in the matrix, is widespread and greatly reduces specificity, leading to false positives and/or overestimation of the analyte concentration (5–7).
The scale of the cross-reactivity problem can be illustrated with a study involving a group of 11,000 affinity-purified, mono-specific monoclonal antibodies (8). Only 531 produced a single band on a Western blot, indicating that 95% bound to ‘non-target’ proteins. Some of these results may be due to protein isoforms, cleaved proteins, or post-translational modifications, but the majority of the low specificity was probably due to cross reactivity.
Thus, testing an antibody for cross-reactivity with closely related proteins is a critical validation experiment. We will look into this in detail in a companion article. The most straightforward solution to avoiding cross-reactivity is careful choice of antibodies. In general, monoclonal antibodies (mAb), which recognize a single epitope, provide high specificity at the expense of sensitivity, since only one antibody molecule can bind to the antigen. Polyclonal antibodies (pAb) provide higher sensitivity since multiple antibodies may bind to a single antigen molecule, but may be more cross-reactive since the epitope is less well defined. Where possible, a mAb should be chosen as the primary antibody (e.g. for capture) to establish high assay specificity, and a pAb can be used as a detection reagent.
Miniaturization eases the pain of sample and reagent consumption
The ability to do more with less is at a premium. More data needs to be generated, more quickly, and for multiple analytes from single samples. These can include small volumes of precious samples such as vitreous and aqueous humor, tears, and synovial fluid that are expensive and difficult to source, e.g. samples from pediatric patients or patients with rare diseases. There is also the desire to fulfill the requirements of the 3R’s (Replace, Reduce, Refine) in animal studies, which includes the need to combine microsampling with sensitive assay methods that consume minimal sample (9). Added to that, there is a demand to use only minimal amounts of expensive reagents such as lead compounds in early drug development.
Miniaturizing immunoassays to the nanoliter scale reduces reagent and sample consumption, making it possible, for example, to perform ‘one mouse, one PK’ studies that minimize animal use and biological variation in preclinical studies and maximize data quality. This approach can lead to 50–80% reductions in animal use and substantial cost savings. The same level of miniaturization can also mean great savings on precious drug candidates prepared in small scale for preclinical and earlier clinical trials – for example, using 1/20th of the capture antibodies and 1/100th of the detection antibodies is possible.
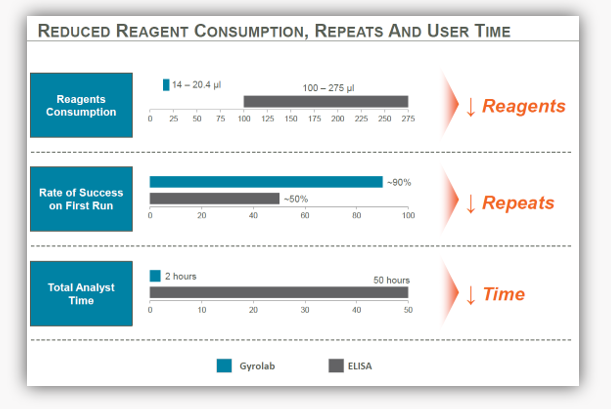
Figure 1. An example of how transferring from a manual or semi-automated ELISA onto an automated platform that generates high precision results using miniaturized immunoassays can reduce reagent consumption, repeats and user time.
Avoiding sample dilution and repeats by staying in range
Sample dilution may be necessary to avoid interference and should be minimized to maintain sensitivity, but it can also be a challenge to dilute samples to ensure that they are in range for the assay to avoid assay repeats. Extensive sample dilution can result in errors, and assay repeats take time and effort. Repeats may also be necessary if the method is not robust, or has a low precision such that outliers are common. The solution to these challenges is to ensure that the method has high precision and a broad analytical range.
Robust automation saves time
One of the greatest challenges in immunoassays is the rapid and efficient generation of high quality data to enable effective data-driven decision making to meet demanding time lines. This relies on choosing a platform that enables you to quickly develop and validate immunoassays that also involve short assay run times to boost productivity. Automation will minimize hands-on time, freeing up scientists for more important tasks such as data analysis. Automation combined with the generation of high precision data also supports the effective use of Design of Experiments (DOE), an approach that can accelerate method development to higher levels.
A miniaturized flow-through immunoassay system that improves your workflow
As we have seen, the challenges of immunoassays can be met with careful choice of reagents and evaluation of assays early in development. The Gyrolab® platform transforms immunoassay performance and productivity by enabling the rapid development of methods that deliver high precision data from small amounts of sample using minimal amounts of precious reagent. As a result, the Gyrolab system has made ‘one mouse, one PK’ a reality. This is possible due to microfluidics in the CD that processes samples in individual microstructures in parallel, which eliminates cross talk and edge effects. The microstructures provide automated volume definition, which removes the influence of manual pipetting error. Added to that the flow-through technology minimizes matrix effects, and the automation and rapid turnaround saves valuable time. Advanced visual tools, such as Gyrolab Viewer software, help in effective reagent screening and quality control of data.
You can find out more about how Gyrolab system has been used to deliver high precision data with minimal consumption of sample and reagents by downloading a case study on the use of Gyrolab system for preclinical PK analysis.
References:
- Interference with clinical laboratory analyses. Kroll MH & Elin RJ. Clin. Chem. 40(11 Pt 1), 1996–2005 (1994).
- Interferences in Immunoassay. Tate, J Greg Ward G. Clin Biochem Rev 2004; 105-120
- Analytical error and interference in immunoassay: minimizing risk. Sturgeon CM & Viljoen A. Ann Clin Biochem 2011; 48: 418–432. DOI: 10.1258/acb.2011.011073
- Interference in immunoassays to support therapeutic antibody development in preclinical and clinical studies. Schwickart et al Bioanalysis (2014) 6(14), 1939–1951
- Antibody array analysis of labelled proteomes: how should we control specificity? Holm A, et al. New Biotechnol 2012, 29:8-585.
- Opportunities for sensitive plasma proteome analysis. Landegren U, et al. Anal Chem 2012, 84:1824-1830.
- Specificity and selectivity evaluations of ligand binding assay of protein therapeutics against concomitant drugs and related endogenous proteins. Jean Lee & Hongjin AAPS J. 2007 May 25;9(2):E164-70.
- Toward next generation plasma profiling via heat-induced epitope retrieval and array-based assays. Schwenk JM, et al. Mol Cell Proteomics. 2010 Nov;9(11):2497-507. doi: 10.1074/mcp.M110.001560. Epub 2010 Aug 3.
- One mouse, one pharmacokinetic profile: quantitative whole blood serial sampling for biotherapeutics. Joyce AP, et al. Pharm Res. 2014 Jul;31(7):1823-33.